Welcome to the Cui Lab
We are a young research group at the Guangzhou National Laboratory. If you are interested in working with us, please see more information on Positions .
Our research team is dedicated to pioneering spatially resolved multi-omics methodologies for delineating the spatiotemporal regulation of cellular fate specification during respiratory organogenesis and disease pathogenesis.
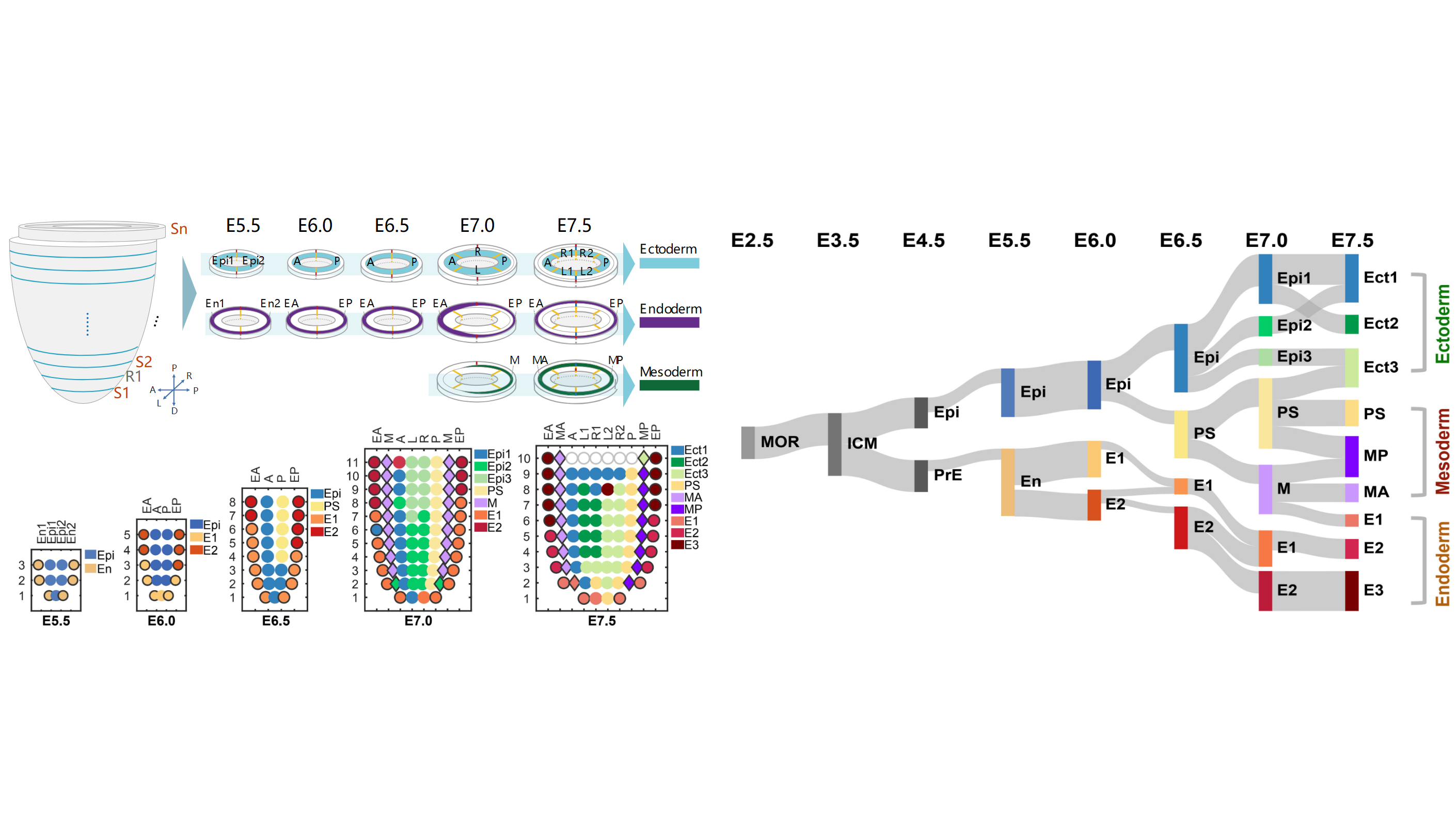
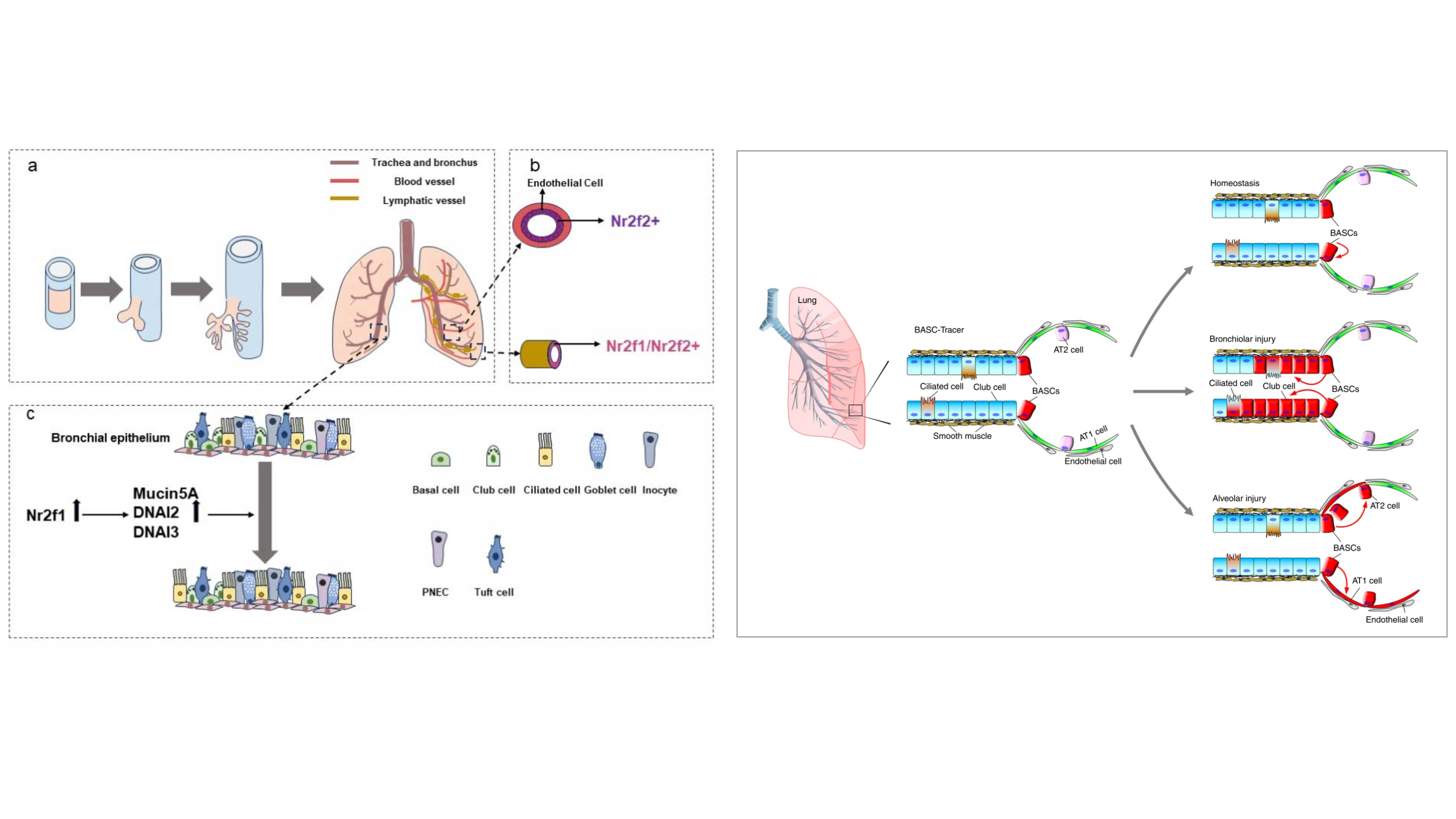
Although organogenesis requires precise spatiotemporal orchestration of developmental programs and carcinogenesis emerges as their dysregulated counterpart through subversion of conserved molecular pathways, these processes share conserved dependencies on proliferative capacity, cellular plasticity, and adaptive microenvironmental niches that drive cellular state transitions. Through methodological breakthroughs, we have developed automated single-cell multi-omics platforms that synergistically integrate transcriptomic profiling, epigenomic mapping, and spatial resolution technologies. These technological innovations have enabled two major breakthroughs: 1. Systematic elucidation of molecular trajectory dynamics during gastrulation and three-dimensional regulatory architectures in organogenesis; 2. Comprehensive characterization of cellular phenotypes and their molecular signatures across pulmonary morphogenesis and neoplastic transformation. Our principal investigators have led multiple collaborative initiatives, making significant contributions as corresponding or first authors in high-impact journals such as Nature, Nature Genetics, and Cell Reports. These achievements have been recognized through nationally prestigious awards, including the ‘Top Ten Breakthroughs in Life Sciences’ and ‘Top Ten Advances in Bioinformatics’ in China.”
Building upon these achievements, our research agenda is dedicated to developing next-generation high-resolution spatial multi-omics platforms that will enable us to elucidate the molecular mechanisms governing cell-cell interaction during early organogenesis while concurrently characterizing immune ecosystem dynamics within tumor microenvironments. By integrating single-cell and spatial omics data, we aim to advance our understanding of both normal developmental processes and pathological transformations at unprecedented spatiotemporal resolution.